Review Article
Surface Plasmon Resonance technology to assess biological interactions

Silvia Bartollino*, Alessandro Medoro, Donatella Mignogna, Erika di Zazzo and Bruno Moncharmont
Department of Medicine and Health Sciences “V. Tiberio”, University of Molise, Italy
*Address for Correspondence: Silvia Bartollino, PhD, Department of Medicine and Health Sciences “V. Tiberio”, University of Molise, Via F. De Sanctis, 86100 Campobasso, Italy, Tel: +39 0874404886; Email: [email protected]
Dates: Submitted: 15 July 2017; Approved: 24 August 2017; Published: 25 August 2017
How to cite this article: Bartollino S, Medoro A, Mignogna D, di Zazzo E, Moncharmont B. Surface Plasmon Resonance technology to assess biological interactions. Insights Biol Med. 2017; 1: 039-044. DOI: 10.29328/journal.ibm.1001005
Copyright License: © 2017 Bartollino S, et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Keywords: SPR; Sensorgram; Ligand; Analyte; Biological interactions
Abstract
Molecular interactions between proteins or between proteins and small molecules are pivotal events for selective binding of biological structures and, consequentially, for their correct function. In this scenario, the evaluation of kinetic parameters, characterizing a molecular interactions, is considered a crucial event to reveal the nature of binding processes.
The focus on peculiar forces involved in the molecular recognition represents an opportunity to explore biological interactions in real time, and to develop a number of innovative biotechnological methods for diagnosis and/or therapy.
Currently, optical biosensors, offering an increasingly effective technology to detect in real time molecular binding, are usually composed by a detector, a sensor surface and a sample delivery system: only definite substances, which are able to interact specifically with the biological part, lead to an optical or electrical signal of the physical transducer.
In this review we want to highlight the exponentially-growing interest of Surface Plasmon Resonance (SPR) based optical biosensors for molecular binding analysis in different research fields.
Biosensors in Biological Interactions
Surface Plasmon Resonance (SPR)-based optical biosensors offer an increasingly effective technology to detect molecular interactions in real time [1,2].
In the last decade, the use of biosensors is enormously increased [3], for molecular recognition in an increasing number of different research fields [4-9]: a fundamental advantage is that biomolecular interactions, ranging from the millimolar to the nanomolar range, can be detected and monitored in real time. The binding of the analyte-free flowing in solution- to the ligand -anchored to the surface of a sensor chip- can be followed because of producing a change in the refractive index, due to a change in mass concentration on the thin surface: this latter is usually composed by a gold layer and the change in the mass value is converted to resonance or response units (RU) in real time, creating a typical plot, called sensorgram. In this way, this detection method doesn’t require any ligand labeled with a radioactive isotope or with a fluorophore. The method is not invasive and it can measure the amount of binding complex formed, even in crude preparations [8,10].
From optical biosensors data, it is possible to calculate the equilibrium affinity constant (KD) which is an important determinant of biological processes, and even more interestingly the association (Kon) and dissociation (Koff) rate constants for biomolecular interaction. Moreover, another advantage in the use of biosensors is that they can be coupled with mass spectrometers, representing an even more powerful technology not necessitating any pre-labeling of molecules, thus avoiding time-and moneyconsuming steps [7]. Furthermore, the amount of material for a binding experiment is very low: typically few micrograms of both ligand and analyte are required [4].
The two commercial instruments that have promoted the optical biosensor technique are the BIAcore (GE-Healthcare Life Sciences, Chicago, USA) using SPR technology, and the IAsys (Affinity Sensors, Cambridge, UK) which is based on a Resonant Mirror (RM) platform. They share two peculiar features: an optically coupled sensor surface with various coatings (for ligand immobilization), and an evanescence wave transducer, which is sensitive to changes of refractive index value of the fluidic compartment (sensing layer), and keeps contact with the sensor layer. The IAsys measurements include stirring of analyte solution above a sensor in the bottom of a cuvette, whereas the BIAcore measurements are carried out under a continuous analyte flow over the sensor surface mounted in a microfluidic flow cell, called Integrated Microfluidic Cartridge (IFC). This peculiar feature allows the accurate measurement of the amount of the analyte-ligand complex during the binding cycle. In the microfluidic system, the valves switch between the running buffer and sample with high precision, in order to deliver the sample as a defined fluidic part with minimum dispersion between sample and buffer [11].
The flow cell configuration allows the buffer to flow continuously over the sensor surface, reducing the need for time-consuming washing steps, when analyte is exchanged for buffer, and minimizing the amount of analyte used for the binding experiment. Furthermore, a continuous flow system guarantees the ligand exposition to a constant analyte concentration during the experiment.
Binding and dissociation events, producing an increase or decrease of molecular mass on the sensor surface, are directly correlated to the refractive index changes at the sensor/bulk phase interface [12], providing a method to monitor kinetic parameters of the binding reactions, with a growing interest for medical chemistry [13].
SPR Technology
SPR technology, as introduced above, is a real-time and label-free method used to investigate binding kinetic constants and affinity data of small molecules (KD 10-8-10-4 M), antibodies (KD 10-12-10-7 M) or protein fragments (KD 10-6-10-3 M) [1-3,14,15].
It can be performed on a biospecific surface composed of a thin gold layer of a sensor chip, which generates physical conditions to induce SPR and which separates two dielectric compartment. The ligand is immobilized on a gold support (approx. 50 nm thick) deposited on a glass part, through chemical modification of the gold film, and the solution containing the analyte is then injected over this modified surface in a continuous flow rate.
Typical reaction times of less than 30 min are required for the immobilization process. The sensor chip typically used in BIAcore system, that employs the Kretschmann configuration [16,17], is a dextran matrix, composed of a thioalkane linker layer (~100 nm thick), that creates an hydrophilic environment for different chemical methods of ligand immobilization, which usually require an amount of ligand immobilized (about 10,000 RU) that corresponds to approximately ≈70 fmol/mm2, and for binding process [18]. The activation of the gold layer and the application of a specific coat avoid non-specific binding of biomolecules to the activated surface [19]. Furthermore, the dextran matrix allows both ligand immobilization with minimal conformational distortion and maximum access of the binding epitopes of ligand and analyte. Suggested Matrix density or thickness by molecular weight of ligand guarantees the number and the accessibility of binding sites, which are optimized on the sensor surface during the analyte injection on the sensor chip surface [12,20]. The selectivity is achieved by coating the sensor surface with a specific ligand, which can be immobilized by direct covalent coupling or noncovalent capture by a covalently immobilized anchor. This step is crucial to ensure a stable attachment of the ligand to the sensor surface, preserving the binding properties of the ligand.
There are many different sensor surfaces, but the most common immobilization procedure is to coat the surface with the carboxymethyldextran (CMD) matrix [10]. This water-based procedure produces a carboxymethyldextran-modified sensor surface, with the formation of N-hydroxysuccinimide esters from a fraction of the carboxyl groups of the carboxymethyldextran matrix reacting with Nhydroxysuccinimide and N-ethyl-N’-(dimethylaminopropyl) carbodiimide hydrochloride. For binding to this solid-phase the ligand is injected over this chemically-modified surface in a solution of low ionic strength with a pH value 1 point below the isoelectric point of the protein. In this way the ligand is captured in the matrix by electrostatic attraction forces allowing a simultaneous reaction with the active residues, which are subsequently transformed into amides - by ethanolamine addition- to complete the immobilization procedure [21].
Analysis of optical parameter modified by analyte interaction to the immobilized ligand is obtained by application of the Fresnel model to SPR data, typically taken by an instrument in a prism based configuration [22]. This response can be detected and quantified as change in the refractivity (refractivity/Response Unit) [20], producing a sensorgram, which is a plot representing the binding between the tethered ligand and the analyte.
Kon and Koff kinetic parameters are measured by subsequent injections of different analyte concentrations on the immobilized ligand, after regeneration steps between each injection. By non-linear regression analysis, it is possible to calculate the isotherm of the binding step and to measure the equilibrium KD of the ligand-analyte complex by plotting binding responses at steady state against different analyte concentrations. Data analysis provides information on the molecular interaction and on the stoichiometry of the complex.
SPR is based on Total Internal Reflection (TIR) phenomenon. As a light beam propagates into a medium of higher refractive index (the glass of the sensor chip in the BIAcore platform) and keeps contact with a medium of lower refractive index (the aqueous sample solution), it generates an evanescent wave into the lower refractive index medium. This wave has some electric field components directed in different orientations during the insertion into the lower refractive index medium and it interacts with mobile electrons in the gold layer at the surface of the glass.
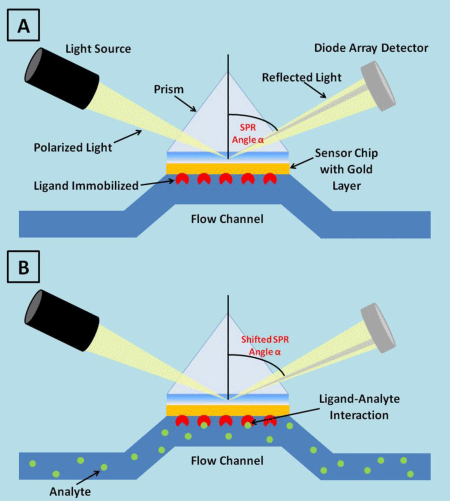
In this fluidic system, the analytes captured by the immobilized ligand over the sensor chip generate a mass accumulation complex on the thin layer, which induces a variation in the resonance angle. SPR causes a reduction in the intensity of reflected light at a specific angle of reflection: it varies with the refractive index close to the surface, on the opposite side from the reflected light (the sample side in BIAcore). In BIAcore platform a wedge-shaped diode of monochromatic light is commonly used. Each angle of the reflected beam strikes the detector at a different point, and a diode array detector (DAD) continuously records the position of reduced light intensity and monitors the SPR angle α [23], as shown in figure 1.
Figure 1: SPR technology detects changes in SPR angle value, resulting in a refractive index change, due to immobilized ligand and analyte complex formation.
The light is not absorbed by the sample and the energy is dissipated through SPR in the gold layer. When the analyte binds to the ligand immobilized on the sensor surface, the refractive index changes, due to the modiβication in solute concentration. The angle of incidence required to create the SPR phenomenon is so altered and this modiβication is measured as a response signal: a resonance unit corresponds to a change of this resonance angle, which arises by a change of the surface density (1 RU=0.01 mDegree = 1 pg of protein per mm2). The change in the value of minimum reβlectivity angle, Surface Plasmon Resonance technology to assess biological interactionsPublished: August 25, 2017042called SPR angle, can be determined by varying the incidence angle and recording the reβlected light intensity during the biological binding reactions between biomolecules. The SPR angle is directly proportional to the amount of the analyte binding to the sensor layer, because it generates a positive SPR signal. The SPR angle change is reported as resonance units (RU), where 1000 RU correspond to an angle change of ~ 0.1 degree. For most proteins, binding of ~ 1 ng of protein/mm² of sensor surface is required to generate a signal change of 1000 RU.
Discussion
Since 1990, SPR technology became a standard biophysical method to perform real-time binding experiments and to evaluate kinetics parameter crucial to investigate biological interaction process. So far, SPR sensors are widely used for biomolecular studies [24,25], in biomedical, environmental and pharmaceutical applications. Data availability on association and dissociation rate, stability of the ligand-analyte complex, lack of labelling requirements, low material consumption, rapidity and versatility of the procedure make SPR technology an excellent platform for high-throughput screening methods, particularly for screening of small molecules at an early stage of drug discovery investigation.
The wide number of applications for SPR biosensor platforms indicates the power of this technology, both in terms of variety of ligands and analytes that can be used, and of sample types (serum, cell lysates and a huge number of buffers). Furthermore, appropriate data analysis of sensorgrams can also shed light onto binding biological process which are more elaborate respect to a 1:1 binding model event or that can happen through conformational changing [26,27]. Examples of areas exploiting SPR technology are food analysis [28], immunogenicity [29,30], proteomics [31], drug discovery [32], epitope mapping investigation [33], “epitope binning” studies for vaccine investigation [34] and ligand βishing [35].
Conclusions
The knowledge of structural biological changes associated due to biomolecular interactions represents the milestone to assess and discover biological process.
Currently new insights are achieved by using SPR technology to measure kinetic parameters (rates of complex association and dissociation) driving biochemical and biological reactions, such as afβinity and speciβicity of molecular interactions, establishing an “afβinity ranking” in which it is possible to discriminate weak binders, stable binders or no binders [36].
A new frontier in this βield is the realization of a miniaturized and multichannel, portable SPR devices [37].
References
- Pattnaik P, Surface Plasmon resonance: applications in understanding receptor-ligand interaction. Appl. Biochem. Biotechnol. 2005; 126: 79-92. Ref.: https://goo.gl/19DKNS
- Lee TH, Hirst DJ, Aguilar MI. New insights into the molecular mechanisms of biomembrane structural changes and interactions by optical biosensor technology. Biochim Biophys Acta. 2015; 1848: 1868-1885. Ref.: https://goo.gl/dNYmGq
- Douzi B. Protein-Protein Interactions: Surface Plasmon Resonance. Methods Mol Biol. 2017; 1615: 257-275. Ref.: https://goo.gl/XNJuYP
- Cooper MA, Optical biosensors in drug discovery. Nat Rev Drug Discov. 2002; 1: 515-528. Ref.: https://goo.gl/Xe1CPP
- Shin HJ, Lee H, Park JD, Hyun HC, Sohn HO, et al. Kinetics of binding of LPS to recombinant CD14, TLR4, and MD-2 proteins. Mol Cells. 2007; 24: 119-124. Ref.: https://goo.gl/Ftf2bb
- Cannon MJ, Papalia GA, Navratilova I, Fisher RJ, Roberts LR, et al. Comparative analyses of a small molecule/enzyme interaction by multiple users of Biacore technology, Anal Biochem. 2004; 330: 98-113. Ref.: https://goo.gl/qF14p8
- Fabini E, Danielson UH. Monitoring drug-serum protein interactions for early ADME prediction through Surface Plasmon Resonance technology. J Pharm Biomed Anal. 2017; 114: 188-194. Ref.: https://goo.gl/je1GRc
- Leonard P, Hearty S, Ma H, O’Kennedy R. Measuring Protein-Protein Interactions Using Biacore. Methods Mol Biol. 2017; 1485: 339-354. Ref.: https://goo.gl/vimT1j
- Nguyen HH, Park J, Kang S, Kim M. Surface plasmon resonance: a versatile technique for biosensor applications. Sensors (Basel). 2015; 15: 10481-10510. Ref.: https://goo.gl/HA8ZYn
- Baird CL, Myszka DG. Current and emerging commercial optical biosensors. J Mol Recognit. 2001; 14: 261-268. Ref.: https://goo.gl/pCDDbj
- Biacore Life Sciences nd. Accessed. 2017. Ref.: https://goo.gl/uGRToc
- Keusgen M. Biosensors: new approaches in drug discovery. Naturwissenschaften. 2002; 89: 433-444. Ref.: https://goo.gl/UHcx9g
- Copeland RA. Drug-target interaction kinetics: underutilized in drug optimization? Future Med Chem. 2016; 8: 2173-2175. Ref.: https://goo.gl/yYfexS
- Ferlini C, Bartollino S, Cicchilliti L, Penci R, Raspaglio G, et al. 471 POSTER Functional assessment of Bcl-2 disordered loop through plasmon surface resonance technology. Eur J Cancer Suppl. 2006; 4: 144.
- Ferlini C, Cicchillitti L, Raspaglio G, Bartollino S, Cimitan S, et al. Paclitaxel Directly Binds to Bcl-2 and Functionally Mimics Activity of Nur77, Cancer Res. 2009; 69: 6906-6914. Ref.: https://goo.gl/RoGG2f
- Jason-Moller L, Murphy M, Bruno J. Overview of Biacore Systems and Their Applications. Curr Protoc Protein Sci. 2006. Ref.: https://goo.gl/4DXqXp
- Rich RL, Papalia GA, Flynn PJ, Furneisen J, Quinn J, et al. Myszka, A global benchmark study using affinity-based biosensors. Anal Biochem. 2009; 386: 194-216. Ref.: https://goo.gl/A5onva
- Ligand immobilization using thiol-disulphide exchange. (n.d.). Accessed. 2017. Ref.: https://goo.gl/9sydnU
- Livnat Levanon N, Vigonsky E, Lewinson O. Real time measurements of membrane protein:receptor interactions using Surface Plasmon Resonance (SPR). J Vis Exp. 2014. Ref.: https://goo.gl/diZfAw
- Jönsson U, Fägerstam L, Ivarsson B, Johnsson B, Karlsson R, et al. Real-time biospecific interaction analysis using surface plasmon resonance and a sensor chip technology. Biotechniques. 1991; 11: 620-627. Ref.: https://goo.gl/aAhdYp
- Johnsson B, Löfås S, Lindquist G. Immobilization of proteins to a carboxymethyldextran-modified gold surface for biospecific interaction analysis in surface plasmon resonance sensors. Anal Biochem. 1991; 198: 268-277. Ref.: https://goo.gl/q9JMrA
- Peterson AW, Halter M, Plant AL, Elliott JT. Surface plasmon resonance microscopy: Achieving a Quantitative optical response. Rev Sci Instrum. 2016; 87: 93703. Ref.: https://goo.gl/eT36G2
- Wilson WD. Tech Sight. Analyzing biomolecular interactions. Science. 2002; 295: 2103-2105. Ref.: https://goo.gl/6wN8hm
- Buijs J, Franklin GC. SPR-MS in functional proteomics. Brief. Funct. Genomic. Proteomic. 2005; 4: 39-47. Ref.: https://goo.gl/vxQMLW
- Nedelkov D, Nelson RW. Analysis of native proteins from biological fluids by biomolecular interaction analysis mass spectrometry (BIA/MS): exploring the limit of detection, identification of non-specific binding and detection of multi-protein complexes. Biosens Bioelectron. 2001; 16: 1071-1078. Ref.: https://goo.gl/cPwUs3
- Geitmann M, Danielson UH. Studies of substrate-induced conformational changes in human cytomegalovirus protease using optical biosensor technology. Anal Biochem. 2004; 332: 203-214. Ref.: https://goo.gl/YBYbDD
- Fabini E, Zambelli B, Mazzei L, Ciurli S, Bertucci C, Surface plasmon resonance and isothermal titration calorimetry to monitor the Ni(II)-dependent binding of Helicobacter pylori NikR to DNA. Anal Bioanal Chem. 2016; 408: 7971-7980. Ref.: https://goo.gl/k6sVxp
- Rebe Raz S, Leontaridou M, Bremer MGEG, Peters R, Weigel S. Development of surface plasmon Resonance-based sensor for detection of silver nanoparticles in food and the environment. Anal Bioanal Chem. 2012; 403: 2843-2850. Ref.: https://goo.gl/dhUWVy
- Fda. HIGHLIGHTS OF PRESCRIBING INFORMATION. @BULLET Indic. Usage Color Cancer. 2009. Ref.: https://goo.gl/jfyGMu
- O’Riordan N, Kilcoyne M, Joshi L, Hickey R. Exploitation of SPR to Investigate the Importance of Glycan Chains in the Interaction between Lactoferrin and Bacteria. Sensors. 2017; 17: 1515. Ref.: https://goo.gl/2v8cbd
- Camperchioli A, Mariani M, Bartollino S, Petrella L, Persico M, et al. Fattorusso, Investigation of the Bcl-2 multimerisation process: Structural and functional implications, Biochim. Biophys. Acta-Mol Cell Res. 2011; 1813: 850-857. Ref.: https://goo.gl/CVwRu5
- Ferlini C, Cicchillitti L, Raspaglio G, Bartollino S, Cimitan S, et al. Paclitaxel Directly Binds to Bcl-2 and Functionally Mimics Activity of Nur77. Cancer Res. 2009; 69: 6906-6914. Ref.: https://goo.gl/QUXySo
- Agadjanyan MG, Zagorski K, Petrushina I, Davtyan H, Kazarian K, et al. Humanized monoclonal antibody armanezumab specific to Nterminus of pathological tau: characterization and therapeutic potency. Mol Neurodegener. 2017; 12: 33. Ref.: https://goo.gl/FFybbR
- Cairns TM, Ditto NT, Lou H, Brooks BD, Atanasiu D, Eisenberg RJ, et al. Global sensing of the antigenic structure of herpes simplex virus gD using high-throughput array-based SPR imaging. PLoS Pathog. 2017; 13. Ref.: https://goo.gl/YfrPhU
- Zhukov A, Schürenberg M, Jansson O, Areskoug D, Buijs J. Integration of surface plasmon resonance with mass spectrometry: automated ligand fishing and sample preparation for MALDI MS using a Biacore 3000 biosensor. J Biomol Tech. 2004; 15: 112-119. Ref.: https://goo.gl/NRB7Yz
- Townsend S, Finlay WJJ, Hearty S, O’Kennedy R. Optimizing recombinant antibody function in SPR immunosensing. The influence of antibody structural format and chip surface chemistry on assay sensitivity. Biosens Bioelectron. 2006; 22: 268-274. Ref.: https://goo.gl/mG1Dxs
- Zhang XL, Liu Y, Fan T, Hu N, Yang Z, et al. Design and Performance of a Portable and Multichannel SPR Device. Sensors (Basel). 2017; 17: 1435. Ref.: https://goo.gl/CpHR5M